Nucleic acid conjugates as self-assembled templates for multivalent presentation of ligands
The ability for mutual recognition is a key characteristic of DNA and RNA. The formation of well-defined complexes proceeds in accordance to the known Watson-Crick base pairing rules. On this basis it is possible to use DNA or RNA as a template to place chemical groups and functional units- if desired- at angstrom precision.
Within the Sonderforschungsbereich 765 we use DNA as a molecular ruler to determine the distance between binding pockets in biological receptors. Here, ligands of biological receptors are covalently attached to an oligonucleotide or peptide nucleic acid oligomer. Only when the spatial arrangement of two or more ligands matches the distance between the binding pockets of the receptors will high bind affinities be obtained.
The principle of spatial screening by chemically modified oligonucleotides was validated for four structurally characterized biological targets. We ascertained the distances between the binding pockets in protein kinases (tandem SH2 domain of Syk kinase) as well as in the estrogen receptor. The investigation of sugar binding proteins (Erythrina Cristigalli lectin, Ricinus Communis agglutinin) showed that bivalency can be explored at distances > 100 Å.
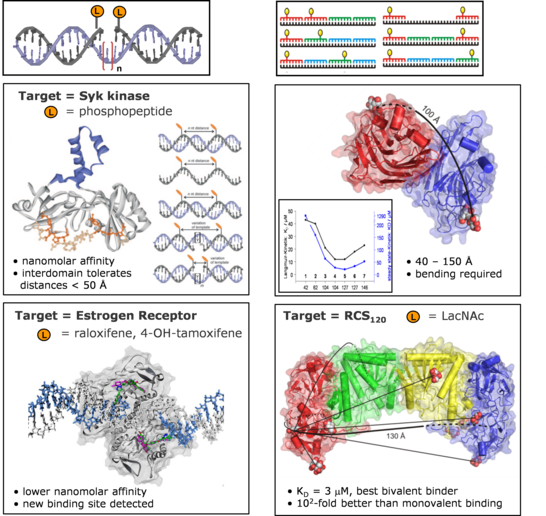
Current efforts focus on the investigation of structurally not characterized target proteins and the spatial screening of receptor aggregates on the surfaces of cells and viruses.